Research overview
Below are four research directions that are currently of great interest to the group. That said, we are always open to exploring new ideas and collaborations.
1. Elucidating disease biology from genetic association studies
Genome-wide association studies (GWAS) have identified hundreds of thousands of genetic variants associated with human disease. Most of these variants lie in non-coding regions of the genome and are generally thought to influence disease risk by regulating the expression of nearby genes. Yet despite extensive efforts, most disease-associated genetic variants remain functionally uncharacterized, with the causal gene unknown in most cases. This has emerged as a major limitation in human genetics, slowing progress toward biological insight and gene-targeted strategy.
A major goal of the lab is to develop integrative statistical approaches to decode variant-to-gene regulation or disease and infer biological mechanisms underlying disease-associated genetic variants. This work will rely on integration of GWAS data with diverse variant-to-gene linking strategies derived from functional genomic datasets, including QTL analyses, CRISPR-based perturbations, enhancer–gene connections from single-cell multiome assays, and chromatin contact maps.
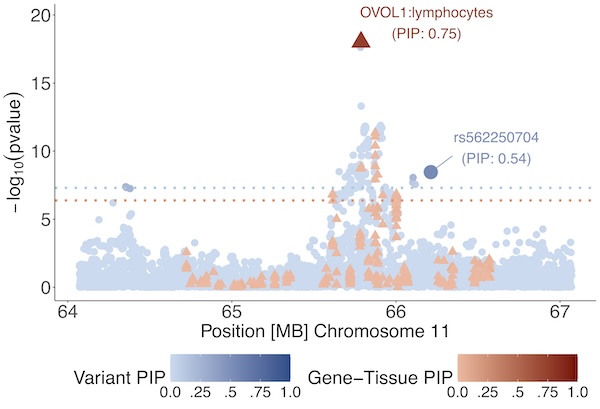
As an example, we developed Tissue-Gene Fine-Mapping (TGFM), a statistical method to uncover the functional mechanisms underlying disease-associated variants by integrating multi-tissue eQTL data with GWAS data (Strober et al., Nature Genetics 2025). TGFM exploits variation in eQTL effects across tissues to pinpoint both the causal gene and its tissue of action at each GWAS locus. Causal gene-tissue pairs identified by TGFM reflected both known biology (for example, TPO–thyroid for hypothyroidism) and biologically plausible findings (for example, SLC20A2–artery aorta for diastolic blood pressure).
A major goal of the lab is to develop integrative statistical approaches to decode variant-to-gene regulation or disease and infer biological mechanisms underlying disease-associated genetic variants. This work will rely on integration of GWAS data with diverse variant-to-gene linking strategies derived from functional genomic datasets, including QTL analyses, CRISPR-based perturbations, enhancer–gene connections from single-cell multiome assays, and chromatin contact maps.
As an example, we developed Tissue-Gene Fine-Mapping (TGFM), a statistical method to uncover the functional mechanisms underlying disease-associated variants by integrating multi-tissue eQTL data with GWAS data (Strober et al., Nature Genetics 2025). TGFM exploits variation in eQTL effects across tissues to pinpoint both the causal gene and its tissue of action at each GWAS locus. Causal gene-tissue pairs identified by TGFM reflected both known biology (for example, TPO–thyroid for hypothyroidism) and biologically plausible findings (for example, SLC20A2–artery aorta for diastolic blood pressure).
2. Modeling genetic effects on gene expression across tissues, cell-types, and contexts
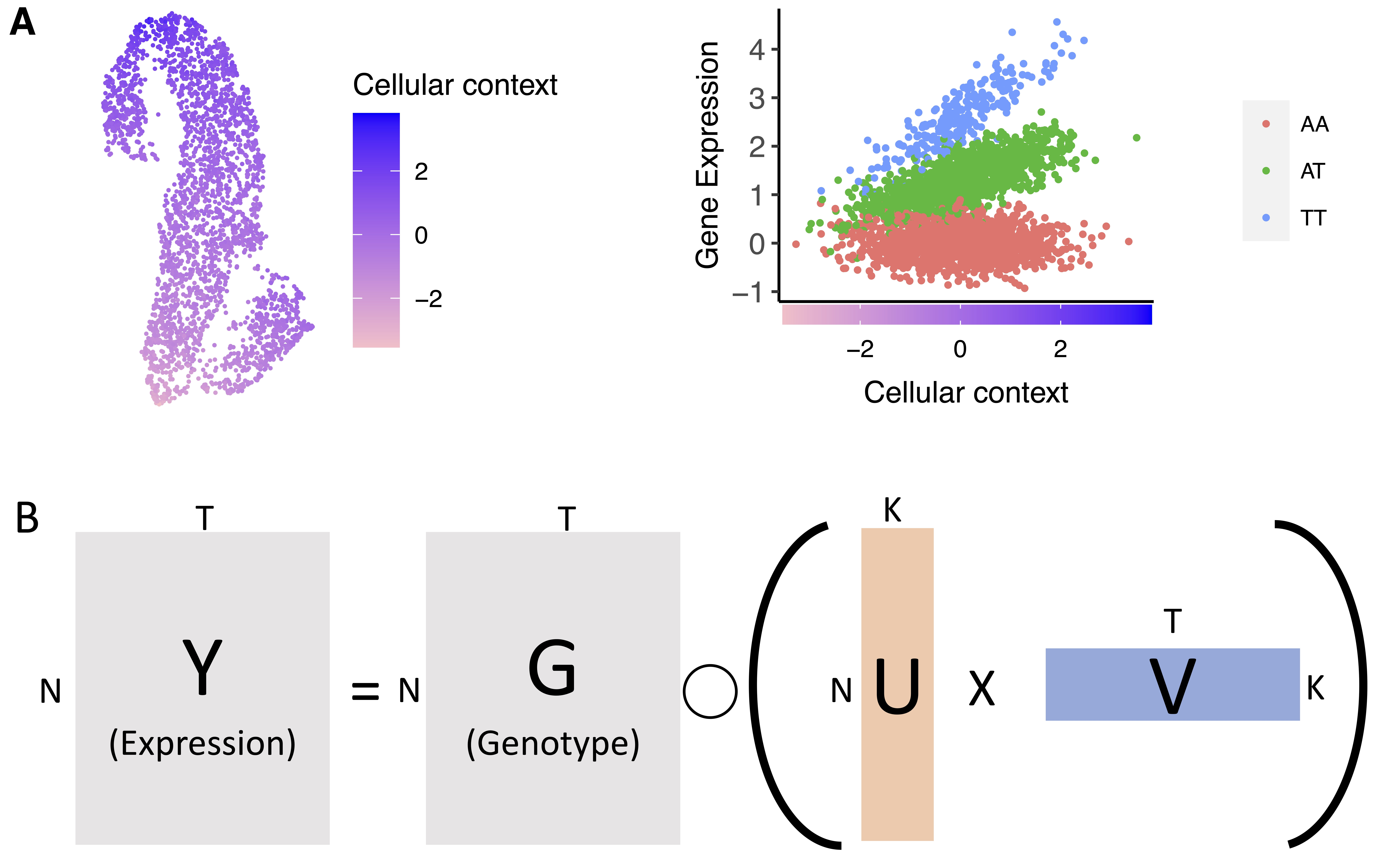
Genetic variants significantly associated with gene expression are known as expression quantitative trait loci (eQTLs). eQTLs are a widely used approach to study the genetic regulation of gene expression and can also be used to infer the biological mechanisms and causal genes underlying disease-associated genetic variants. Dr. Strober has led several projects characterizing how eQTLs vary across tissues and cellular contexts. This includes work with the GTEx consortium, as well as other work demonstrating that eQTLs are highly dynamic statistical associations that change continuously during processes such as cellular differentiation (Strober*, Elorbany*, Rhodes* et al., Science 2019). He also developed a probabilistic model, SURGE, to identify highly context-specific eQTLs from single-cell RNA sequencing using latent-factor models (Strober et al., Genome Biology 2024). Together, these studies demonstrated that genetic regulation of gene expression is highly dynamic and depends on cellular context.
3. Advancing genomically-informed precision medicine for rare diseases
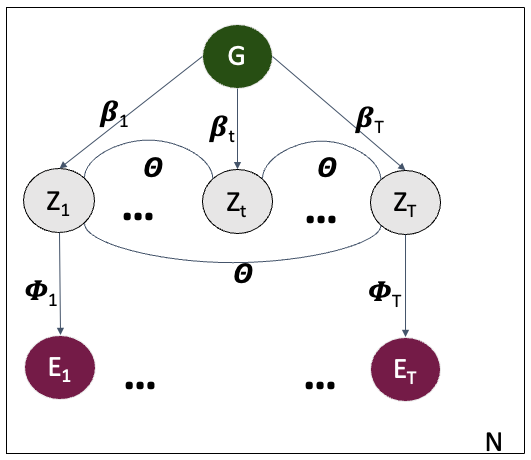
The human genome contains tens of thousands of rare (minor allele frequency <1%) variants, some of which contribute to common and/or rare disease risk. However, determining which of these variants have functional impact or phenotypic consequences remains a major challenge, hindering the diagnosis of rare disease cases. Dr. Strober has developed statistical approaches that leverage individual-level gene expression data to help prioritize functional rare variants (Ferraro*, Strober* et al., Science 2020). This work is motivated by the intuition that if a rare variant has a functional effect, it is likely to perturb the expression levels of nearby genes. More specifically, he developed Watershed, a probabilistic model that integrates genomic annotations (e.g., CADD scores, VEP consequences) with each individual’s transcriptional profile to estimate the posterior probability that a given rare variant has functional consequences.
Looking ahead, we are enthusiastic about continuing this line of work and developing new methods to advance precision medicine for rare disease cases at Boston Children's Hospital. Specifically, we aim to build well-calibrated models that predict disease status or treatment response by integrating clinical, genetic, and multi-omic data. By well-calibrated, we mean that when our model predicts a 70% probability of treatment response, approximately 70% of future patients with that prediction will, in fact, respond. We believe that well-calibrated predictions are essential for adoption of genomically-informed precision medicine into practice.
Looking ahead, we are enthusiastic about continuing this line of work and developing new methods to advance precision medicine for rare disease cases at Boston Children's Hospital. Specifically, we aim to build well-calibrated models that predict disease status or treatment response by integrating clinical, genetic, and multi-omic data. By well-calibrated, we mean that when our model predicts a 70% probability of treatment response, approximately 70% of future patients with that prediction will, in fact, respond. We believe that well-calibrated predictions are essential for adoption of genomically-informed precision medicine into practice.
4. Inferring variant-to-gene links using deep-learning sequence-to-expression models
Many disease loci are thought to have highly context-specific functional mechanisms. For example, recent work found numerous disease loci that overlapped with eQTLs only when expression was measured in endothelial cells exposed to caffeine. This suggests that addressing the missing regulation problem would require population-scale QTL data across countless contexts. We are therefore very interested in exploring scalable alternatives that use sequence-to-expression (S2E) deep learning models in place of QTL data to (1) identify variant-to-gene links across diverse contexts and (2) connect these links to disease to uncover novel disease genes.
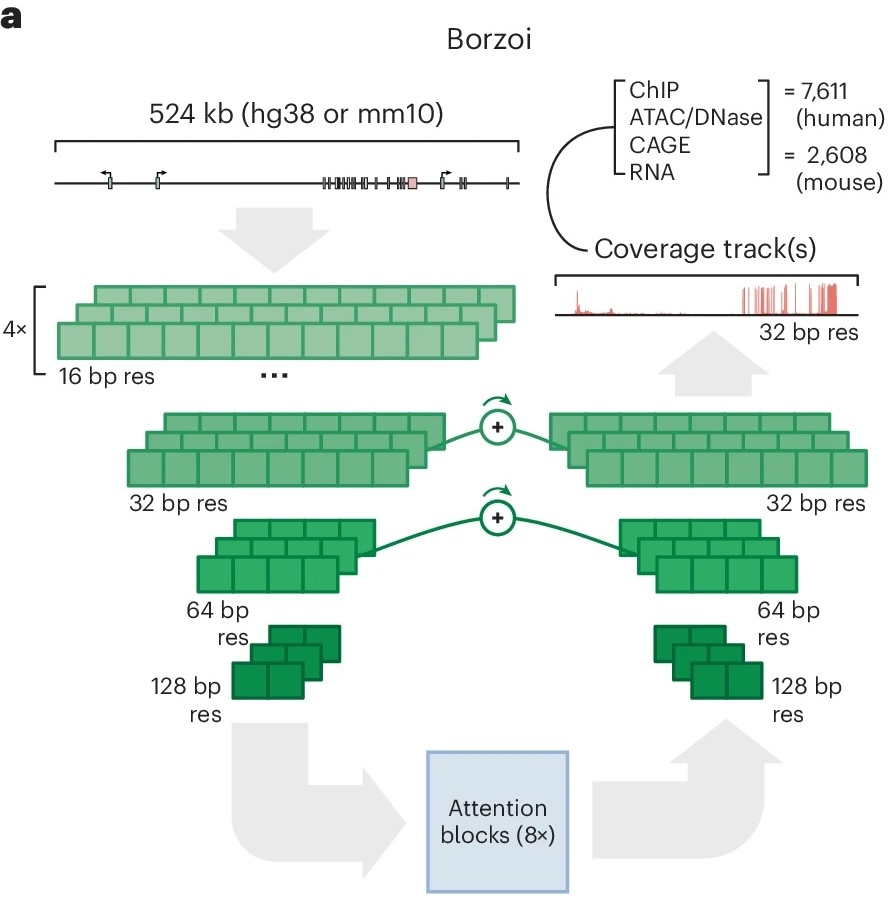
S2E models (e.g., AlphaGenome, Borzoi) leverage deep learning to predict gene expression across tissues, cell types, and contexts directly from DNA sequence. They can also estimate a variant’s effect on a gene by computing the predicted change in expression after substituting the reference with the alternative allele. This provides a complementary alternative to QTL-based estimates. Notably, S2E-predicted effects can be derived without population-scale data, making them particularly valuable for hard-to-assay contexts.
We are currently developing new approaches to integrate S2E predictions with GWAS data to gain insight into the functional mechanisms that underlie disease-associated variants.
S2E models (e.g., AlphaGenome, Borzoi) leverage deep learning to predict gene expression across tissues, cell types, and contexts directly from DNA sequence. They can also estimate a variant’s effect on a gene by computing the predicted change in expression after substituting the reference with the alternative allele. This provides a complementary alternative to QTL-based estimates. Notably, S2E-predicted effects can be derived without population-scale data, making them particularly valuable for hard-to-assay contexts.
We are currently developing new approaches to integrate S2E predictions with GWAS data to gain insight into the functional mechanisms that underlie disease-associated variants.